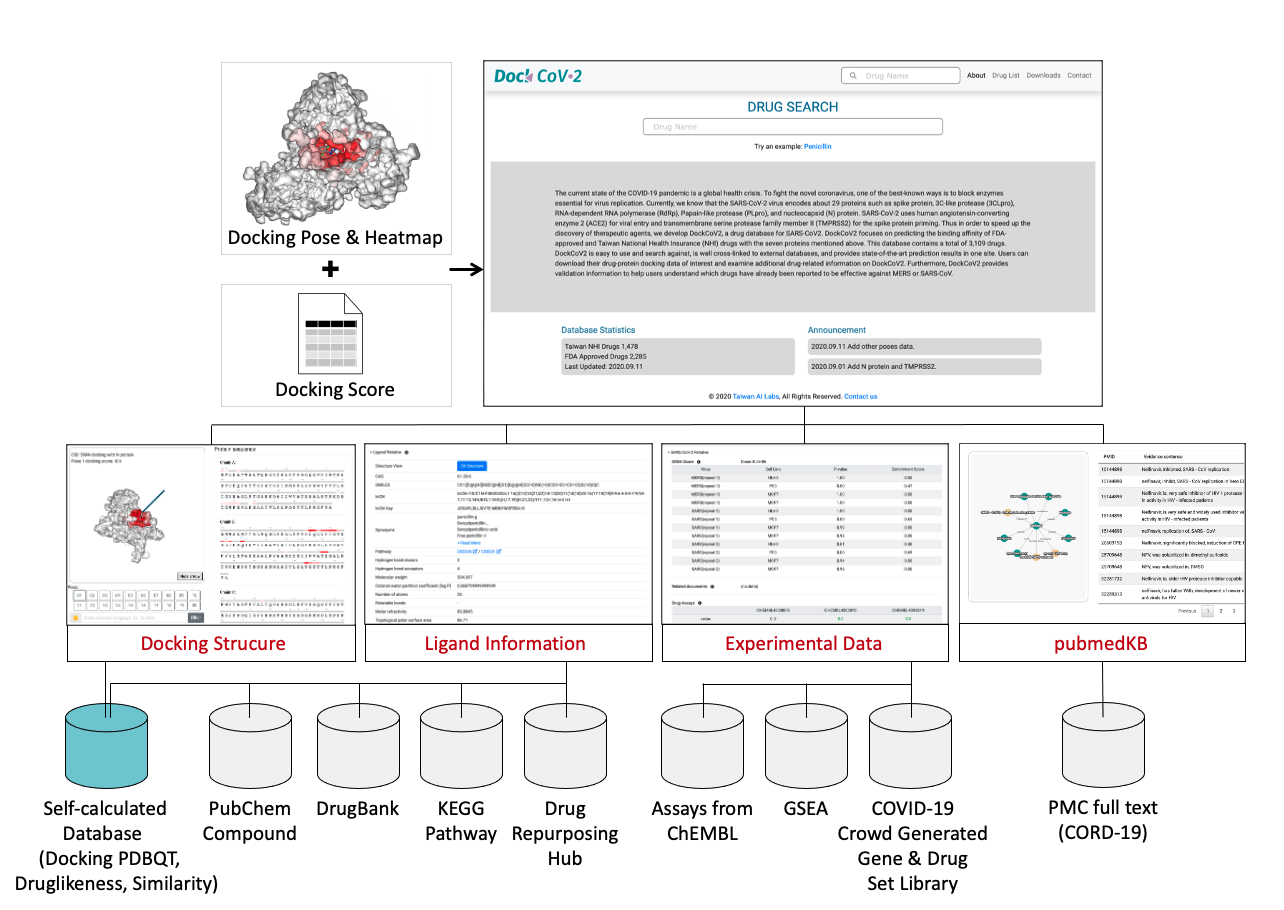
DRUG SEARCH
The current state of the COVID-19 pandemic is a global health crisis. To fight the novel coronavirus, one of the best-known ways is to block enzymes essential for virus replication. Currently, we know that the SARS-CoV-2 virus encodes about 29 proteins such as spike protein, 3C-like protease (3CLpro), RNA-dependent RNA polymerase (RdRp), Papain-like protease (PLpro), and nucleocapsid (N) protein. SARS-CoV-2 uses human angiotensin-converting enzyme 2 (ACE2) for viral entry and transmembrane serine protease family member II (TMPRSS2) for the spike protein priming. Thus in order to speed up the discovery of therapeutic agents, we develop DockCoV2, a drug database for SARS-CoV2. DockCoV2 focuses on predicting the binding affinity of FDA-approved and Taiwan National Health Insurance (NHI) drugs with the seven proteins mentioned above, 5 major SARS-CoV-2 variant proteins and other 67 human proteins, that were identified to be associated with SARS-CoV-2 from GWAS analysis and protein-virus interactions. This database contains a total of 3,548 drugs. DockCoV2 is easy to use and search against, is well cross-linked to external databases, and provides state-of-the-art prediction results in one site. Users can download their drug-protein docking data of interest and examine additional drug-related information on DockCoV2. DockCoV2 also provides validation information to help users understand which drugs have already been reported to be effective against MERS or SARS-CoV. Furthermore, we propose a custom literature-based knowledge graph embedding tool, pubmedKB, for identifying drug and disease relations from published COVID-19-related papers. Specifically, pubmedKB mined over 160 thousand PubMed Central (PMC) full-text literature curated by CORD-19 by applying the state-of-the-art text mining tools from annotation to identification of the drug-disease relations.